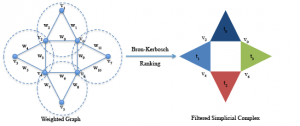
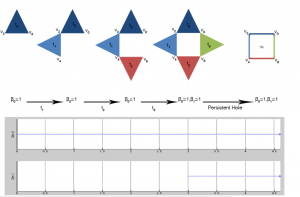
Idiotypic Network
DATASET TYPE: Time-Depending Weighted network.
DATASET DESCRIPTION: The dataset is a collection of temporal networks representing the evolution of the idiotypic network of the mammal immune system. Each vertex in a network represents a class of antibodies. Two vertices are connected if they are immune affine. The weighting function is given by the co-existence coefficient that is:
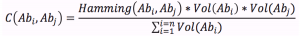
Where, is the Hamming distance between the bit-strings for the antibodies and it expresses the immune affinity between them. represents the concentration of the antibodies .
DATASET ANALYSIS:
- Topological Characterization of Complex Systems: Using Persistent Entropy. Emanuela Merelli, Matteo Rucco, Peter M.A. Sloot & Luca Tesei [2015] Entropy, 17(10), 6872-6892
- Characterization of the idiotypic network through persistent entropy. Matteo Rucco, Filippo Castiglione, Emanuela Merelli & Marco Pettini [2015]. Springer Proceedings in Complexity.
DATASET SOURCE AND DATASET REFERENCE: Massimo Bernaschi and Filippo Castiglione. Design and implementation of an immune system simulator. Computers in Biology and Medicine, 31(5):303–331, 2001.
Epileptic Brain
DATASET TYPE: Multivariate (23 channels) time series
The EEG signals were collected at the Children’s Hospital Boston, and they consist of EEG recordings from pediatric subjects with intractable seizures. Subjects were monitored for up to several days following withdrawal of anti-seizure medication in order to characterize their seizures and assess their candidacy for surgical intervention. We converted the signals into a collection of time-evolving networks by using the correlation coefficient among the signals.
DATASET ANALYSIS:
- A topological approach for multivariate time series characterization: the epilepsy case study Emanuela Merelli, Marco Piangerelli, Matteo Rucco & Daniele Toller [2015] Proceedings of the 9th EAI Conference on Bio-inspired Information and Communications Technologies (BICT 2015)
DATASET SOURCE AND DATASET REFERENCE: http://www.physionet.org
Epidermal Cells
DATASET TYPE: Time-depending multilayer weighted network.
The dataset is a collection of temporal multilayer networks representing the evolution of epidermal cells. Epidermal cells sequentially pass three compartments, named proliferative (pc), differentiated (dc), and stratum corneum (sc). At each time step a network representation of the compartments is given by connecting the cells using both their admissible evolution (i.e., proliferative are connected only with differentiated and differentiated with stratum) and their concentration.
DATASET ANALYSIS:
- jHoles: A Tool for Understanding Biological Complex Networks via Clique Weight Rank Persistent Homology Jacopo Binchi, Emanuela Merelli, Matteo Rucco, Giovanni Petri & Francesco Vaccarino [2014] Electronic Notes in Theoretical Computer Science 306, 5-18.
DATASET SOURCE AND DATASET REFERENCE: Ronald Gieschke and Daniel Serafin. Development of Innovative Drugs via Modeling with MATLAB. Springer, 2013.
Pulmonary Embolism
DATASET TYPE: Dataset formed by categorical and ordinal variables. Each row corresponds to a patient
DATASET DESCRIPTION: A pulmonary embolism is a blockage of the main artery of the lung or one of its branches, frequently fatal. The dataset is formed by 27 diagnostic features of 1,427 patients considered to be at risk of pulmonary embolism enrolled in the Department of Internal and Subintensive Medicine of an Italian National Hospital “Ospedali Riuniti di Ancona”. Patients arrived in the department after a first screening executed by the emergency room.
DATASET ANALYSIS:
- Using topological data analysis for diagnosis pulmonary embolism. Matteo Rucco, et al. Lorenzo Falsetti, Damir Herman, Tanya Petrossian, Emanuela Merelli, Cinzia Nitti & Aldo Salvi [2015]. Journal of Theoretical and Applied Computer Science 9, 1.
- Neural hypernetwork approach for pulmonary embolism diagnosis. Matteo Rucco, Filippo Castiglione, Emanuela Merelli & Marco Pettini [2015]. BMC Research Notes , 8, 1554
- A data-driven clinical prediction rule for pulmonary embolism. Lorenzo Falsetti, Emanuela Merelli, Matteo Rucco, Cinzia Nitti, T. Gentili, M. Pennacchioni & Aldo Salvi [2013]. European Heart Journal 34.suppl 1
DATASET SOURCE AND DATASET REFERENCE: Human clinical variables collected by the department of Sub-intensive medicine of the National Hospital of Ancona and used for the diagnosis of pulmonary embolism.
Falsetti, L. and Merelli, E. and Rucco, M. and Nitti, C. and Pennacchioni, M. and Salvi, A. A data-driven clinical prediction rule for pulmonary embolism. European Heart Journal, The Oxford University Press, 34:243, 2013.
RNA Sequences
DATASET TYPE: Sequences collection of 5s ribosomal RNA and U1 spliceosomal RNA
DATASET DESCRIPTION: suboptimal structures from RNA sequences of 5s rRNA of 120 nucleotides (Rfam accession number RF00001) and three U1 Spliceosomal RNA family with 161 nucleotieds (Rfam accession number X06810.1/261-421), 160 nucleotides (accession number X06809.1/232- 392) and 163 nucleotides (Rfam accession number z11883.1/1496-1656) analyzed using topological data analysis. Besides, to identify structural homology (in biological sense) among species, 63 RNA sequences of 34 species from six family of Archaea namely: Archaeoglobales, Halobacteriales, Methanobacteriales, Methanococcales, Methanomicrobiales and Methanosarcinales are taken from 5S rRNA Database.
DATASET ANALYSIS:
- Persistent Homology Analysis of the RNA Folding Space. Adane Mamuye & Matteo Rucco [2015]. Proceedings of the 9th EAI Conference on Bio-inspired Information and Communications Technologies (BICT 2015)
DATASET SOURCES AND DATASET REFERENCES: Rfam database (http://rfam.xfam.org) and 5s ribosomal RNA database (http://www.man.poznan.pl/5SData/).
- P. Nawrocki, S. W. Burge, A. Bateman, J. Daub, R. Y. Eberhardt, S. R. Eddy, et. al. (2014). Rfam 12.0: updates to the RNA families database. Nucl. Acids Res. 43 (D1): D130-D137.
- Szymanski, M., Barciszewska, M. Z., Barciszewski, J., & Erdmann, V. A. (2000). 5S ribosomal RNA database Y2K. Nucleic Acids Research, 28(1), 166-167.
Direct Current Motors (DC-Motors)
DATASET TYPE: Univariate time series
The signals have been acquired using a 24 bit National Instruments cDAQ data acquisition board (NI-9234) for accelerometer and microphones and a NI-9215 data acquisition board for all the other signals. Sample rate frequency was 51.2 kHz and proper anti-aliasing filters were used. A 10 Hz high-pass filter has been used to remove from acceleration signals the contribution of the rotation around the y axis.
DATASET ANALYSIS:
- Topological classification of small DC motors Matteo Rucco, E. Concettoni, Cristina Cristalli, Andrea Ferrante & Emanuela Merelli [2015] IEEE Proceedings on 1st International Forum on Research and Technologies for Society and Industry Leveraging a better tomorrow (RTSI).
- A new topological entropy-based approach for measuring similarities among piecewise linear functions Matteo Rucco, Rocio Gonzalez-Diaz, Maria-Jose Jimenez, Nieves Atienza, Cristina Cristalli, Enrico Concettoni, Andrea Ferrante & Emanuela Merelli [2015]arXiv: 1512.07613v2
DATASET SOURCE AND REFERENCE:
- E. Concettoni, C. Cristalli, AND S Serafini. Mechanical and electrical quality control tests for small DC motors in production line. In IECON 2012-38th Annual Conference on IEEE Industrial Electronics Society, pages 1883–1887. IEEE, 2012.